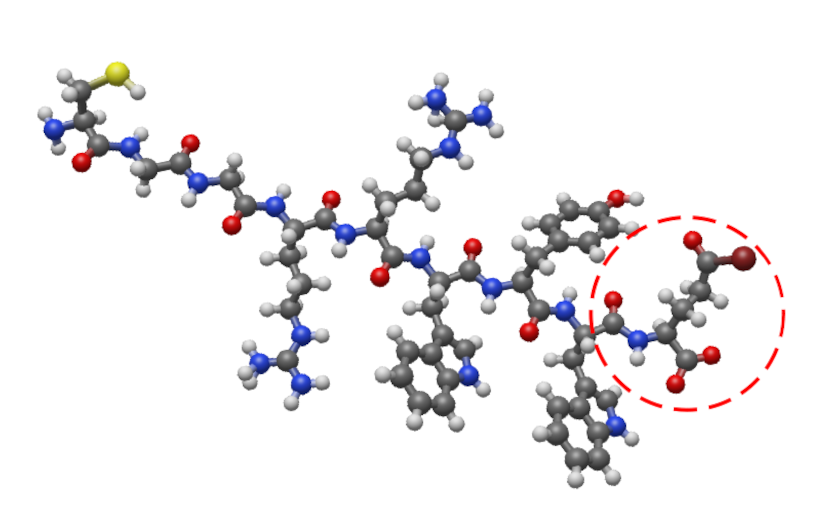
Checking Bonds in Molecular files
In order to create/model polymers that the initial structures are provided with a correct bond description. Unfortunately, due to different reasons, the bonds of the moieties are not correctly described generating conflicts with PySoftK.
Therefore, we have created an auxiliary function called check_bonds located in pysoftk.tool.utils_ob that
perform an automatic check of bonds and reassigne them based on OpenBabel ‘s connect the dots and perceive bond orders
algorithms.
For instance, the following molecule displays an incorrect bond between some atoms (highlighted in the figure by a red circle):
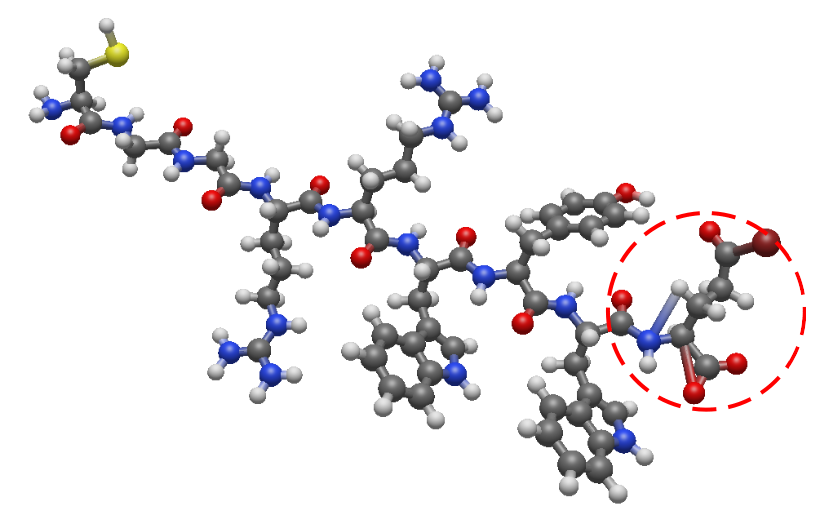
First, we need to import the corresponding modules from PySoftK to allow us to read and join the monomers using RDKit.
import os
from pysoftk.tools.utils_ob import *
Then, the full path of the file containing the molecule needs to be included (as presented in the snippet), and subsequently used in the function check_bonds, as displayed in the following code:
test_file = os.path.join(os.getcwd(), 'rrwtwe_ba_kk_psk.pdb')
check_bond_order(test_file)
Thus, the result of the reconstructed structure (always called with an additional suffix new in the original name) can be shown in the figure below: