Creating diblock Polymers
For building linear diblock copolymers, PySoftK has a specific module (pysoftk.topologies.diblock)
which can be used. The command Db(ma,mb,atom).diblock_copolymer(len_block_A,len_block_B,FF,iter_ff)
is used to generate a diblock copolymer that consists of a block containing len_block_A monomers of ma and a block containing len_block_B monomers of mb.
from rdkit import Chem
from rdkit.Chem import AllChem
mol_1=Chem.MolFromSmiles('BrCOCBr')
mol_2=Chem.MolFromSmiles('[C@H](CBr)(OBr)C')
Then, we can create a polymer with a diblock topology, by using the following commands:
from pysoftk.topologies.diblock import *
di=Db(mol_1,mol_2,"Br").diblock_copolymer(5,7,"MMFF",10)
The new molecular unit (stored in the variable di) can be used to replicate and form a polymer with a given desired number of units (in total 12). The structure can be printed in XYZ format by adding the following lines of code as can be seen in this snipet:
from pysoftk.format_printers.format_mol import *
Fmt(di).xyz_print("diblock.xyz")
By using a common visualization program (such as VMD), the built structure diblock.xyz can be displayed and the result as presented above
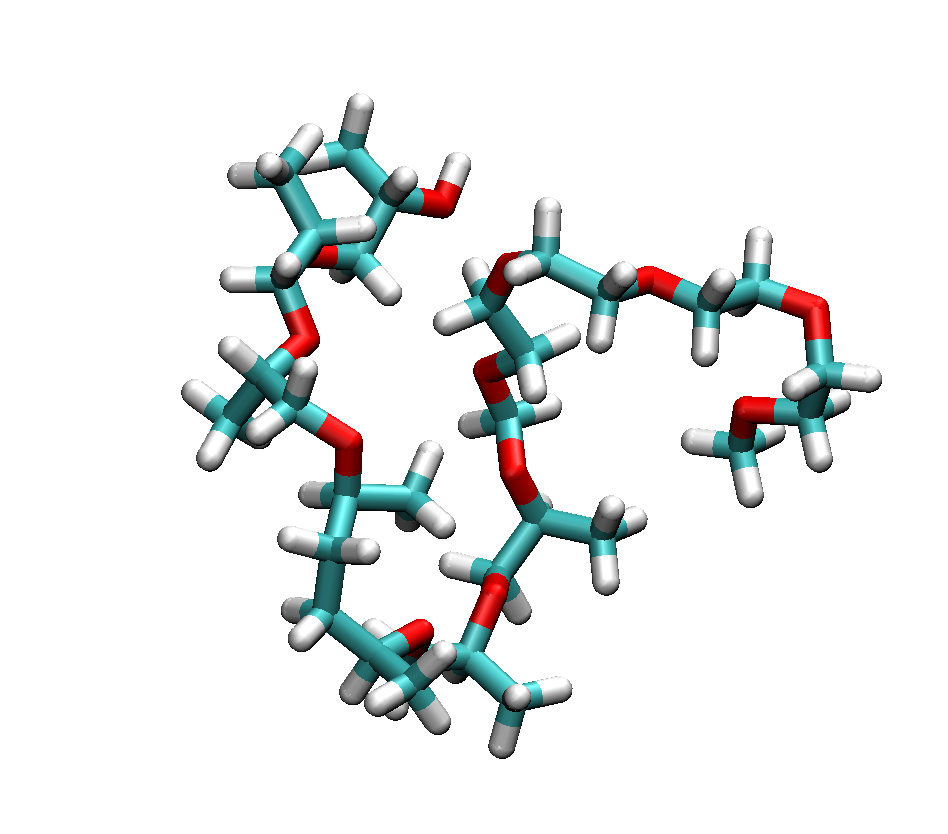
Figure Diblock polymer with 12 units.