Creating new polymers from different modules
Another important functionality of PySoftK is the possibility to create different polymer architectures by combining different modules.
Let’s create a patterned polymer by permuting four monomers using the pysoftk.topologies.diblock function.
First, we need to import the corresponding modules from PySoftK to allow us to read and join the monomers using RDKit.
import rdkit
from rdkit import Chem
from rdkit.Chem import AllChem
from itertools import permutations
from pysoftk.topologies.diblock import *
from pysoftk.linear_polymer.utils import *
Then, we can create the alphabetic permutations and define the monomers whose SMILES codes have been inputted using RDKit :
from pysoftk.topologies.diblock import *
from pysoftk.linear_polymer.utils import *
Once the monomers have been created, the PySoftK function can be used to create the patterned polymer:
string=[''.join(values) for idx, values in enumerate(alph1)]
mols=[Chem.MolFromSmiles('c1cc(oc1Br)Br'),
Chem.MolFromSmiles('c1cc(sc1Br)Br'),
Chem.MolFromSmiles('c1(ccc(cc1)Br)Br'),
Chem.MolFromSmiles('c1(ccc(nc1)Br)Br')]
patt=Pt(string[0], mols, "Br").pattern_block_poly(swap_H=False)
The newly created polymer topology can be used as an initial monomer for futher modelling.
In this case, we use the linear polymer function (pysoftk.linear_polymer.super_monomer)
uses the previous patterned polymer as initial monomer and creates a new linear polymer
of two-unit repetition.
# Linear polymer module
from pysoftk.linear_polymer.linear_polymer import *
from pysoftk.format_printers.format_mol import *
new=Lp(patt,"Br", 2, shift=1.0).linear_polymer("MMFF", 5)
Fmt(new).xyz_print("ABCD_pol.xyz")
The final script is:
import rdkit
from rdkit import Chem
from rdkit.Chem import AllChem
from itertools import permutations
from pysoftk.topologies.diblock import *
from pysoftk.linear_polymer.utils import *
# Creating permutations for 4 different units
alph1=permutations(["A", "B", "C", "D"], 4)
string=[''.join(values) for idx, values in enumerate(alph1)]
mols=[Chem.MolFromSmiles('c1cc(oc1Br)Br'),
Chem.MolFromSmiles('c1cc(sc1Br)Br'),
Chem.MolFromSmiles('c1(ccc(cc1)Br)Br'),
Chem.MolFromSmiles('c1(ccc(nc1)Br)Br')]
patt=Pt(string[0], mols, "Br").pattern_block_poly(swap_H=False)
# Linear polymer module
from pysoftk.linear_polymer.linear_polymer import *
from pysoftk.format_printers.format_mol import *
new=Lp(patt,"Br", 2, shift=1.0).linear_polymer("MMFF", 5)
Fmt(new).xyz_print("ABCD_pol.xyz")
By using common visualization software (such as VMD), the built structure combined_func.py can be displayed as shown:
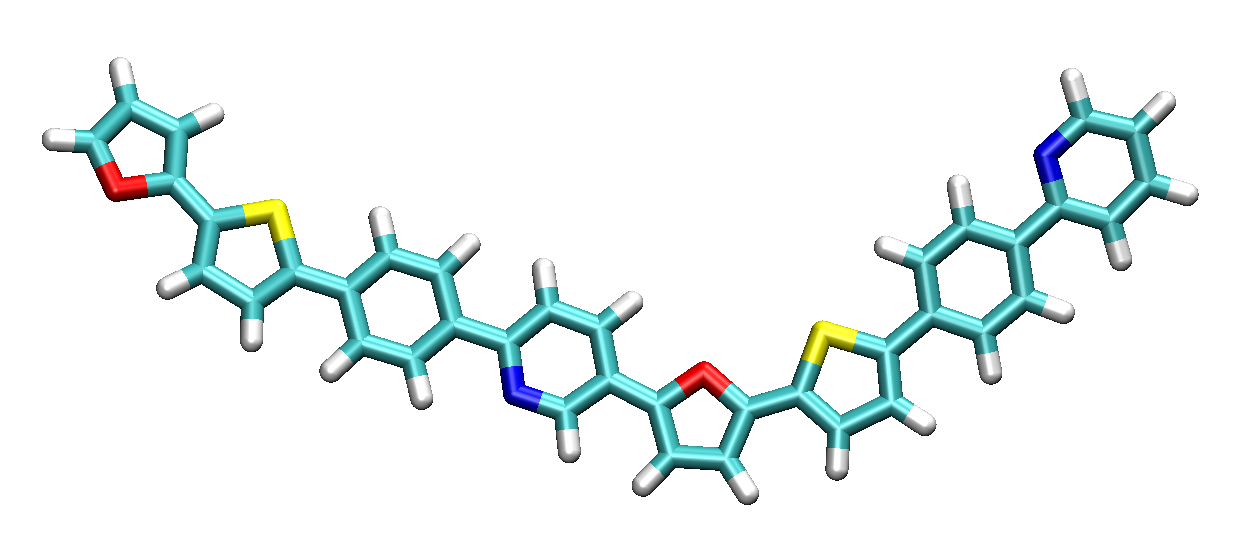
Creating a patterned-linear polymer using different modules of PySoftK.