From single molecules to polymers using PySoftK
PySoftK enables to create Polymers from a single monomer. To do so, the user needs to highligthing the positions where the merging process will take place, using a place-holder atom.
An example is in this section presented using a Styrene molecule. By using a common visualization program (such as VMD), the functionalized molecule is presented below:
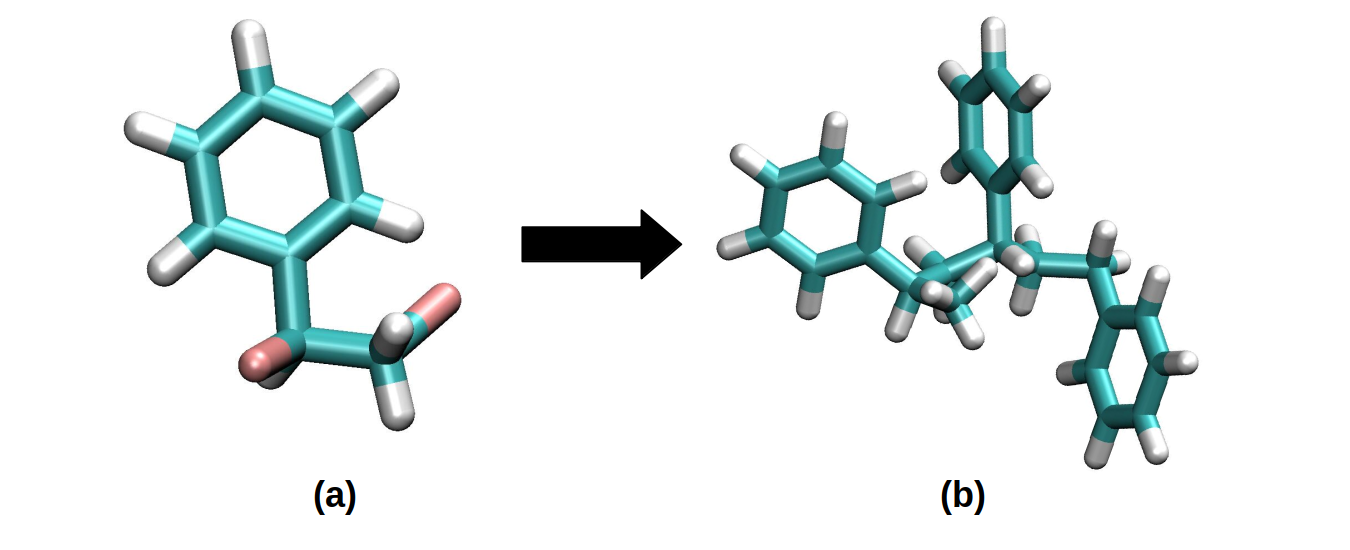
(a) Represents a functionalized single Styrene molecule in which two Bromine atoms are used as place-holders for bonding formation. (b) A Styrene polymer formed with a 3-unit repetition.
The process to build these kind of polymers is presented in this snapshot:
from rdkit import Chem
from rdkit.Chem import AllChem
from pysoftk.linear_polymer.linear_polymer import *
from pysoftk.format_printers.format_mol import *
a=Chem.MolFromSmiles('c1c([C@@H](CBr)Br)cccc1')
#Embedding is needed for being parsed as a pysoftk.object
AllChem.EmbedMolecule(a)
new=Lp(a,"Br",2,shift=1.25).linear_polymer("MMFF",350)
Fmt(new).xyz_print("styrene_pol.xyz")
The Styrene molecule (a) is initially declared using SMILES format. The molecule has been embedded using one the methods available in RDKit and then
parsed to pysoftk.linear_polymer.super_linear_polymer to create an initial polymer structure.
Stereochemistry issues
In some cases, the used monomer exhibits complicated three-dimensional structures which are sometimes not well described using SMILES format. As an example, the following molecule:
a=Chem.MolFromSmiles('[C@H](CBr)(OBr)C')
Produces the following error, when directly parse to PySoftK:
ValueError: Bad Conformer Id
This indicates that the molecule has many possible three-dimensional representations. This problem can be solved, by creating a first set of three- dimensional coordinates in the following way:
from rdkit import Chem
from rdkit.Chem import AllChem
from pysoftk.linear_polymer.linear_polymer import *
from pysoftk.format_printers.format_mol import *
#Creatin a 3-D representation by inactivating the isomericSmiles option from RDKit.
a=Chem.MolFromSmiles('[C@H](CBr)(OBr)C')
b=Chem.MolToSmiles(a, isomericSmiles=False)
c=Chem.MolFromSmiles(b)
# Original Embedding
AllChem.EmbedMolecule(c)
new=Lp(c,"Br",4,shift=1.25).linear_polymer("MMFF",350)
Fmt(new).xyz_print("solved.xyz")
Enabling the creation of the corresponding 4-unit polymer.