Creating a Polymer
Let’s try to create our first polymer by merging two similar monomers using the pysoftk.linear_polymer.super_monomer function.
First, we need to import the corresponding modules from PySoftK to allow us to read and join the monomers using RDKit.
from rdkit import Chem
from rdkit.Chem import AllChem
from pysoftk.linear_polymer.super_monomer import *
from pysoftk.format_printers.format_mol import *
Then, we can create the monomers whose SMILES codes have been inputted using RDKit :
mol_1=Chem.MolFromSmiles('c1cc(sc1Br)Br')
mol_2=Chem.MolFromSmiles('c1(ccc(cc1)Br)Br')
Once the monomers have been created, the PySoftK function can be used to join them :
a=Sm(mol_1,mol_2,"Br").monomer()
The newly created polymer topology can be printed in XYZ format using PySoftK function:
Fmt(a).xyz_print("first_monomer.xyz")
The final script is:
from rdkit import Chem
from rdkit.Chem import AllChem
from pysoftk.linear_polymer.super_monomer import *
from pysoftk.format_printers.format_mol import *
mol_1=Chem.MolFromSmiles('c1cc(sc1Br)Br')
mol_2=Chem.MolFromSmiles('c1(ccc(cc1)Br)Br')
a=Sm(mol_1,mol_2,"Br").monomer()
Fmt(a).xyz_print("first_monomer.xyz")
By using common visualization software (such as VMD), the built structure super_monomer.py can be displayed as shown:
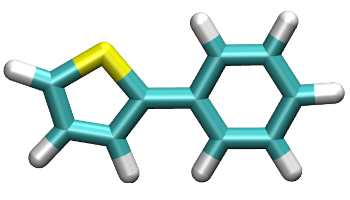
The Thiol and Benzene mioeties have been joined creating a super-monomer that can be further used by PySoftK to create different configurations, topologies and/or investigate torsional angles in a polymer.