Creating chains of Polymers
Let’s try to create our first polymer by merging two similar molecules using the pysoftk.linear_polymer.super_linear_polymer function.
First, we need to import the corresponding modules from PySoftK allowing us to merge the initial molecules (i.e create a super-monomer) and read them using RDKit
from rdkit import Chem
from rdkit.Chem import AllChem
from pysoftk.linear_polymer.super_monomer import *
from pysoftk.linear_polymer.linear_polymer import *
from pysoftk.format_printers.format_mol import *
Then, the molecules are created (using SMILES format) and read using the following lines of code:
mol_1=Chem.MolFromSmiles('c1cc(sc1Br)Br')
mol_2=Chem.MolFromSmiles('c1cc(sc1Br)Br')
Once the initial molecular units are defined, we can create a polymer (i.e a super-monomer in PySoftK language) as indicated in the following lines:
a=Sm(mol_1,mol_2,"Br")
k=a.mon_to_poly()
The new molecular unit (stored in the variable k) can be used to replicate and form a polymer with a given desired number of units (in this case 3) and printed in XYZ format, as can be seen in this code snipet:
new=Lp(k,"Br",3,shift=1.0).linear_polymer()
Fmt(new).xyz_print("polymer_1_2.xyz")
By using a common visualization program (such as VMD), the built structure polymer_1_2.xyz can be displayed and the result as presented above
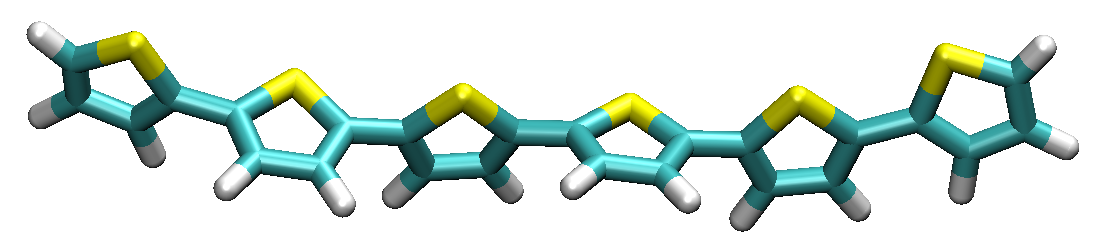
Figure Linear polymer with a 3-unit repetition.